QsarDB predictor service supports DRAGON descriptors
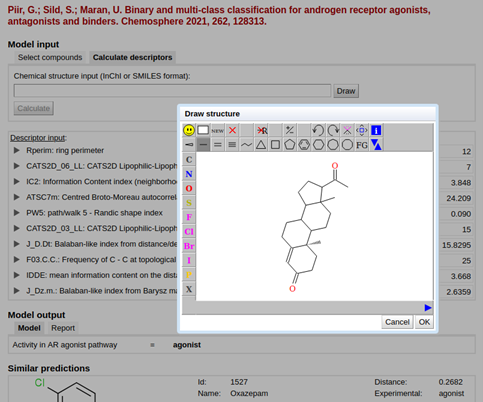
Each model in QsarDB can be used for prediction if the user calculates or determines experimentally the descriptors in the model and enters them in the “QDB Predictor” application [1] or via the REST service [2]. For some models, the prediction service also works directly from the structure. In this case, the QsarDB repository calculates the required molecular descriptors on the fly. Currently, QsarDB supports this functionality for a limited number of models, and they use open source software (Chemistry Development Kit, PaDEL-Descriptor and XlogP3) to calculate molecular descriptors.
However, many QSAR models use commercial software to calculate molecular descriptors. We now took the first step in using a commercial molecular descriptor calculation software and implemented DRAGON 6 to compute molecular descriptors directly from the structure. Thus, all model developers who use DRAGON 6 calculated descriptors in models and bring their model to the QsarDB repository can take the opportunity to make their model predictive directly from the compound structure. An example archive is the QsarDB archive with models for classifying androgen receptor agonists, antagonists, and binders.
At QsarDB we are grateful to Prof. Todeschin for permission to use DRAGON software to calculate molecular descriptors behind the prediction service in the QsarDB repository.
Please contact the QsarDB team (qsardb@chem.ut.ee) for guidance how this functionality can be achieved for your model.
Good prediction!
Your QsarDB team
[1] http://qsardb.org/guidelines/repository/predictor
[2] http://qsardb.org/guidelines/predict
Screenshot from “QDB Predictor” application in structure drawing mode