Compounds
Compounds can be added to the archive manually (one by one) or they can be imported from external files (in bulk). A detailed information about all the fields can be found in the Compounds section for QsarDB data format. It is important to note that every Compound object can hold several structure cargos (e.g. both 2D and 3D representations).
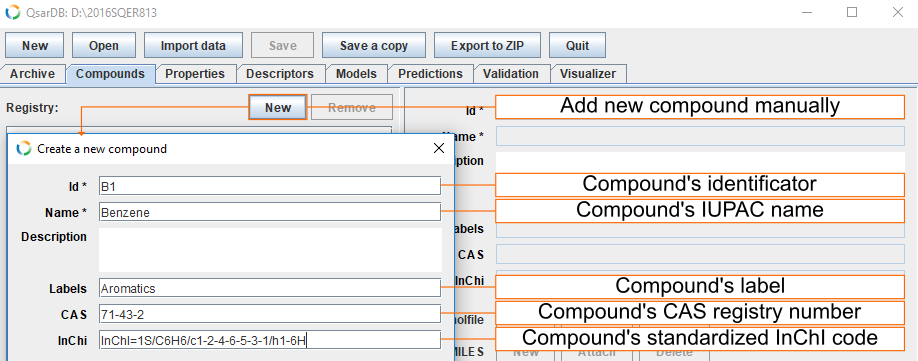
Add compounds manually
Add general information about compound
Button New opens a dialog box, where one can insert attributes that identify the compound(s). The example shows the attributes of the benzene molecule.
- Id is mandatory and, if possible, it should be the same as in the corresponding article. This is the only attribute that cannot be edited later in the QsarDB Editor.
- Name is chemical name and it is mandatory. Recommendation is to give IUPAC name, but common names or names used by the original author are also acceptable.
- Description field (optional) is used for description about compound and for example comes useful when model developer wants to draw attention of model user to the manipulations needed for modelling task.
- Labels can be used to define different sets (optional). Multiple labels should be separated by commas.
- CAS field is used for compound's Chemical Abstract Service registry number (if known).
- InChI field is compound's standard InChI code (strongly recommended).
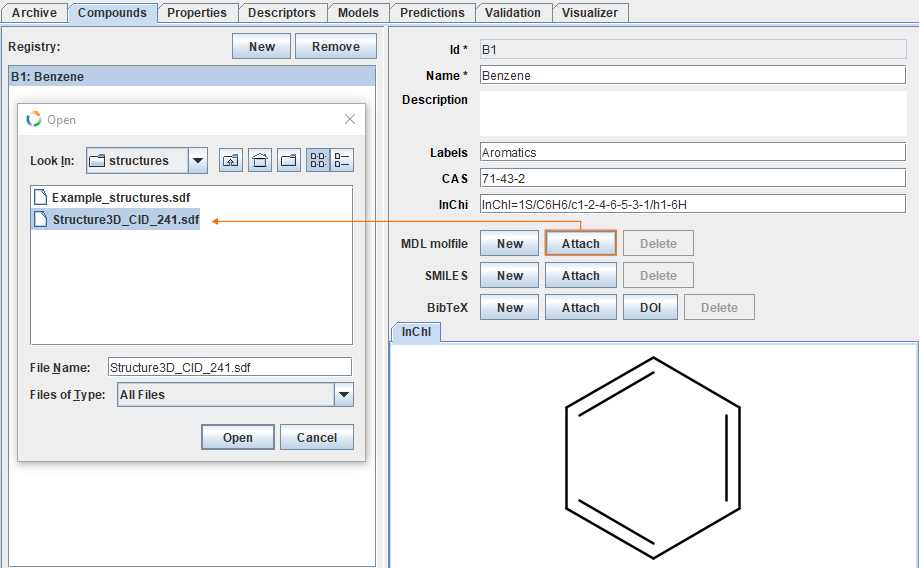
Add MDL molfile
For each compound it is possible to add 3D coordinates used in calculation of descriptors, for example as MDL molfile. Button Attach opens dialog, where one can locate the needed file. For this example, 3D coordinates for benzene can be found here.
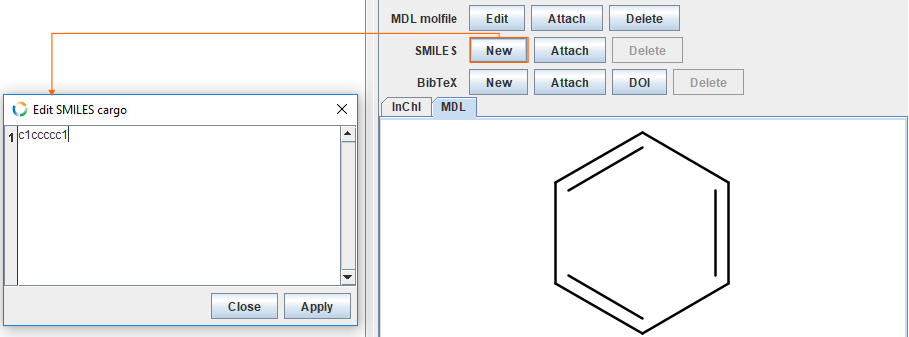
Add SMILES
The SMILES representation for benzene is c1ccccc1. Button New opens text editor, where one can insert SMILES code for the given structure..
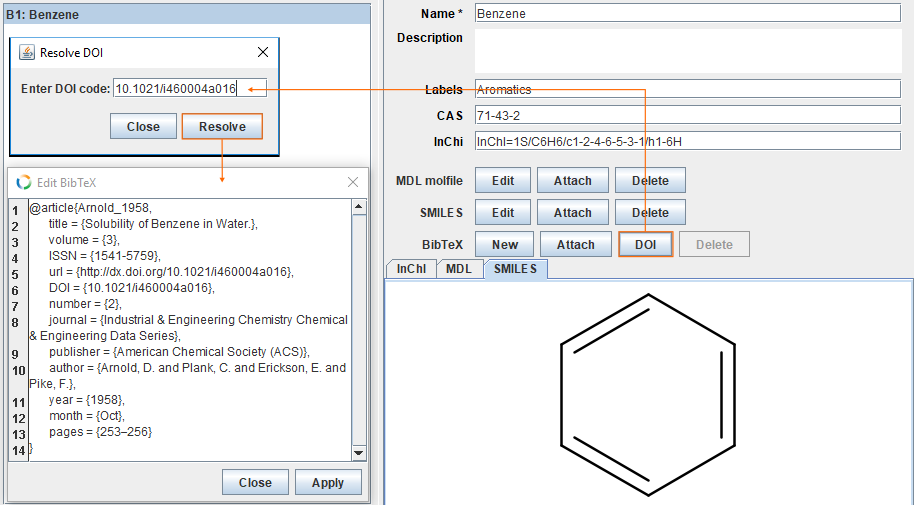
Add reference
It is also possible to add references for each compound. For this example, we are going to use DOI code (10.1021/i460004a016) to download a BibTeX file from the DOI system web services. Button DOI opens dialog for DOI code. Insert DOI code and click Resolve. If DOI code is resolvable, the BibTeX file is downloaded. Check the correctness of the reference and click Apply.
Import compounds from data file
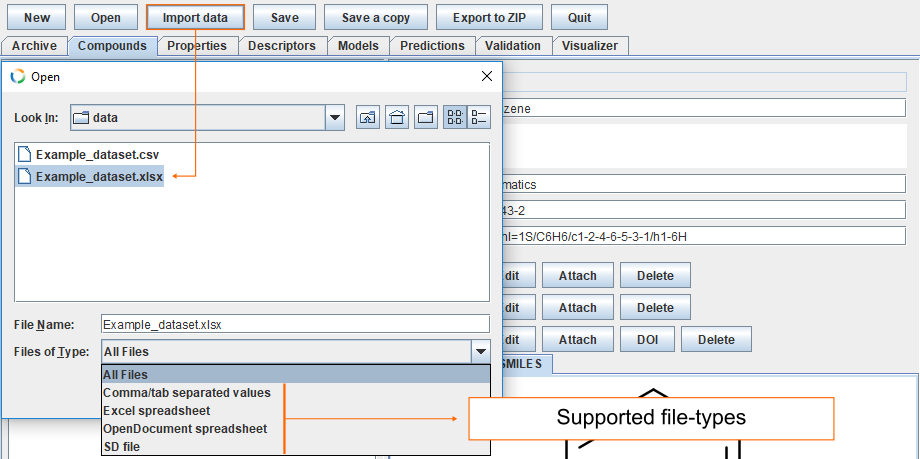
Supported file formats
Usually QSAR datasets deal with multiple compounds and adding them one by one is laborious therefore import from external sources is more practical. The given example uses Excel spreadsheet, (for other file types see supported file formats). Button Import data opens dialog. Find the correct datafile and click Open.
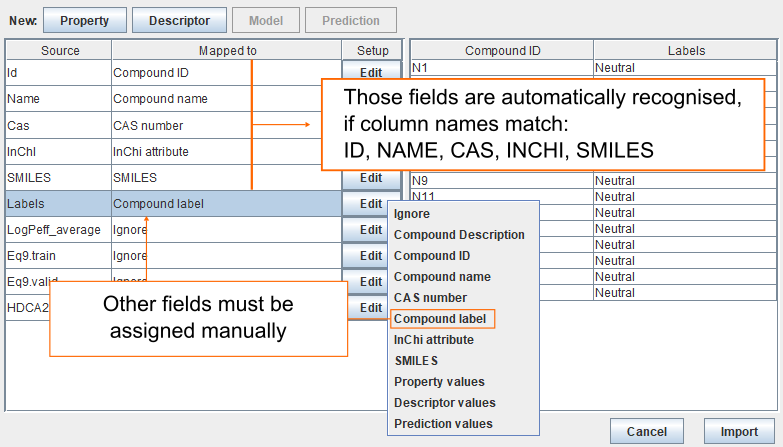
Import compounds from spreadsheet
If column names in the datafile match ID, NAME, CAS, INCHI, SMILES, then they are automatically recognised. Other fields must be assigned manually. To assign column called Labels as Compound label click Edit and select Compound label. Finally, click Import to add all the compounds to the archive.
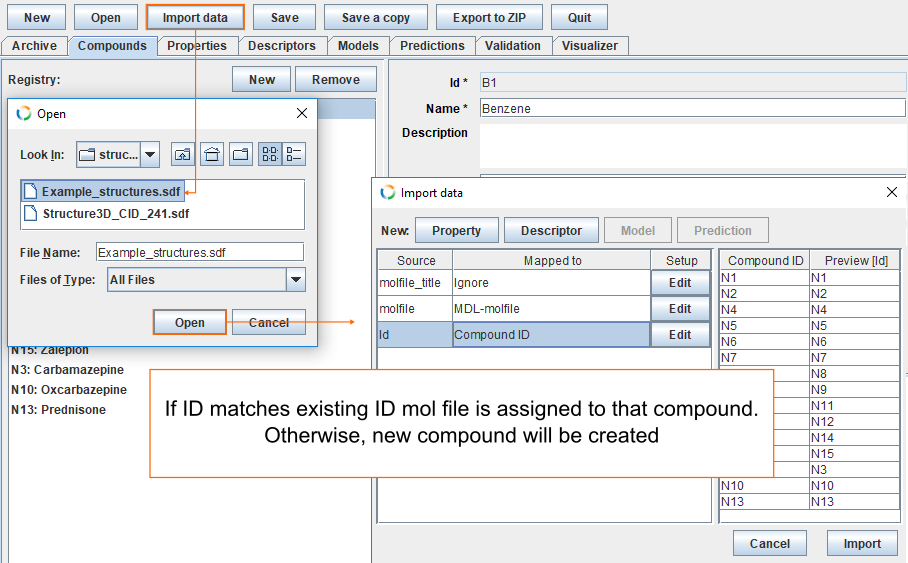
Import compounds from mol-file
To add mol-files to the archive, click Import data, find the file Example_structures.sdf and click Open. If the ID in SDF file matches an existing ID, the mol-file is automatically assigned to that compound. Otherwise, a new compound will be created.
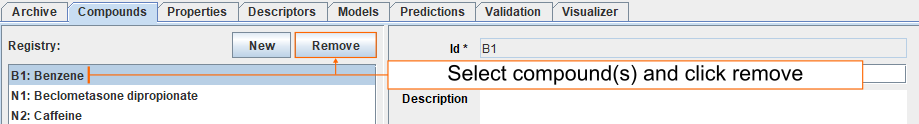
Delete compounds
Select compound(s) you want to delete and click Remove. Multiple compounds can be selected by holding down Shift or Ctrl key.