Step 3: Import data
Import data file
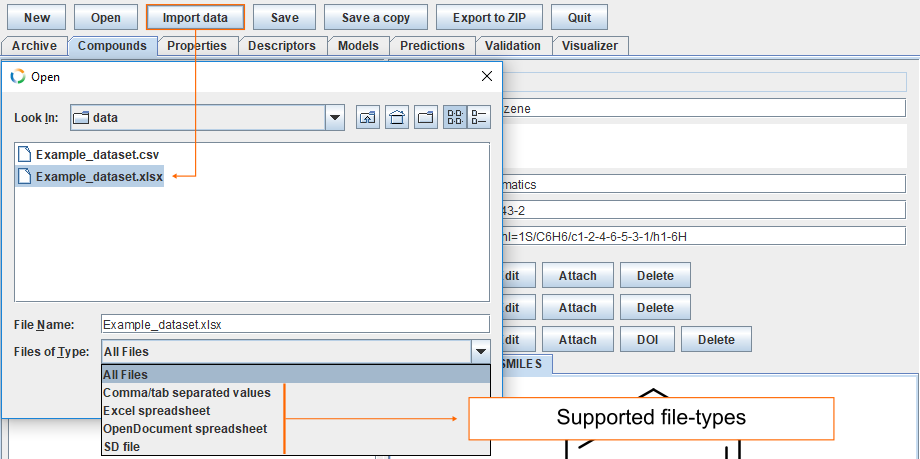
Click Import data and select Example_dataset.xlsx
Map compounds
Since the column names in the datafile match ID, NAME, CAS, INCHI, or SMILES, then they are automatically recognised and automatically mapped. The remaining fields must be assigned manually. To assign the column called Labels as the Compound label, click Edit and then select Compound label.
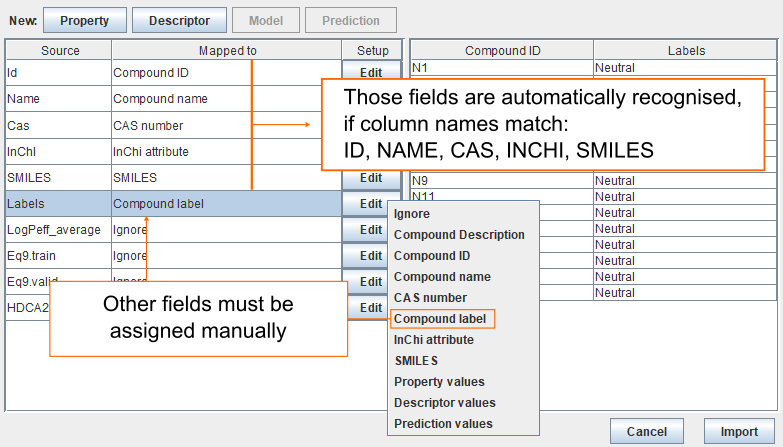
Map properties
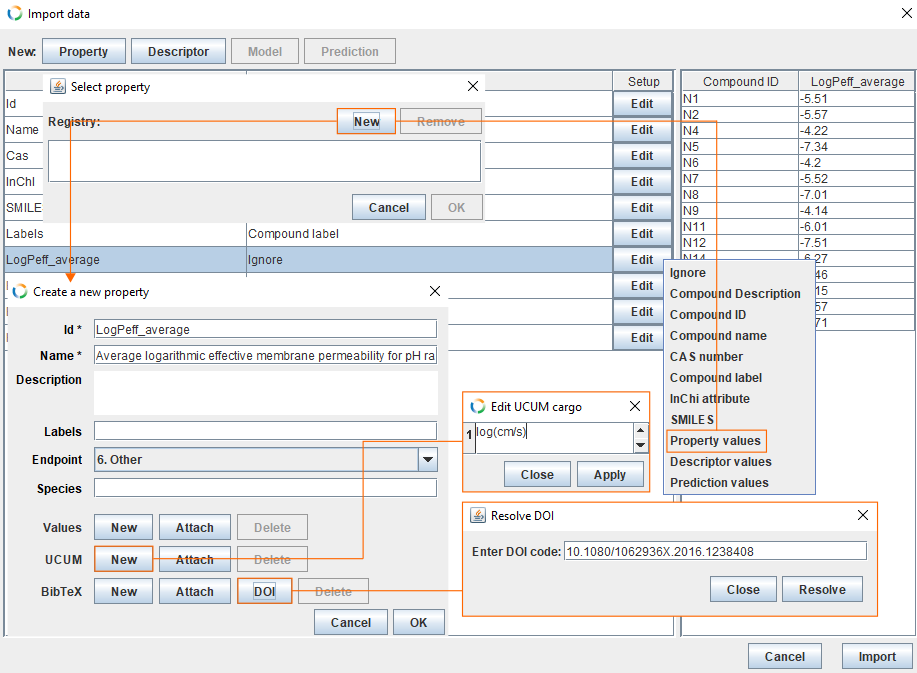
- The column LogPeff_average contains values for property. Click Edit and select Property values.
- This opens a new dialog from where you can select correct property. In this example, we haven't created the property yet. Therefore, click New and fill the fields.
- By default Id field is filled with columns name (LogPeff_average).
- For Name we use following sentence: "Average logarithmic effective membrane permeability for pH range 3 to 9 of neutral compounds".
- For Endpoint choose 5. Toxicokinetics 5.3. Gastrointestinal absorption.
- Add unit by clicking New in the UCUM section, insert log(cm/s) and click Apply.
- Add reference information by clicking DOI in the BibTeX section. Insert 10.1080/1062936X.2016.1238408 and click Resolve. It may take few seconds for the editor to resolve DOI code and download the corresponding BibTeX file. Check the correctness of the reference and click Apply.
- Click OK, select property LogPeff_average and click OK.
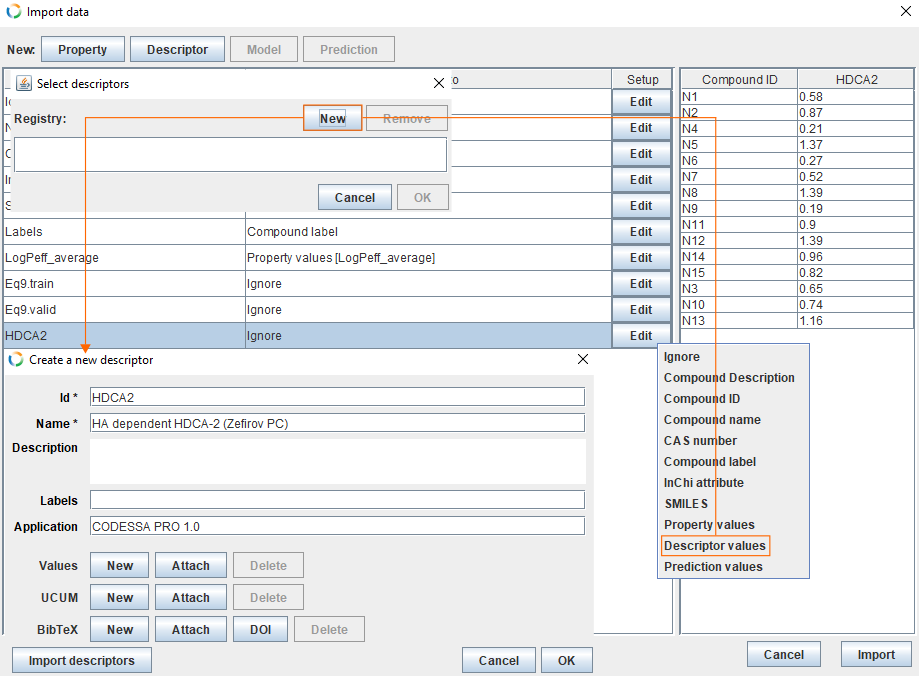
Map descriptors
- The column HDCA2 contains values for the descriptor. Click Edit and select Descriptor values.
- This opens a new dialog from where you can select and correct descriptor. In this example, we haven't created the descriptor yet. Therefore, click New and fill the fields.
- By default Id field is filled with columns name (HDCA2).
- For Name we use descriptor name from the software: "HA dependent HDCA-2 (Zefirov PC)".
- For Application we use: CODESSA PRO 1.0.
- Click OK, select descriptor HDCA2 and click OK.
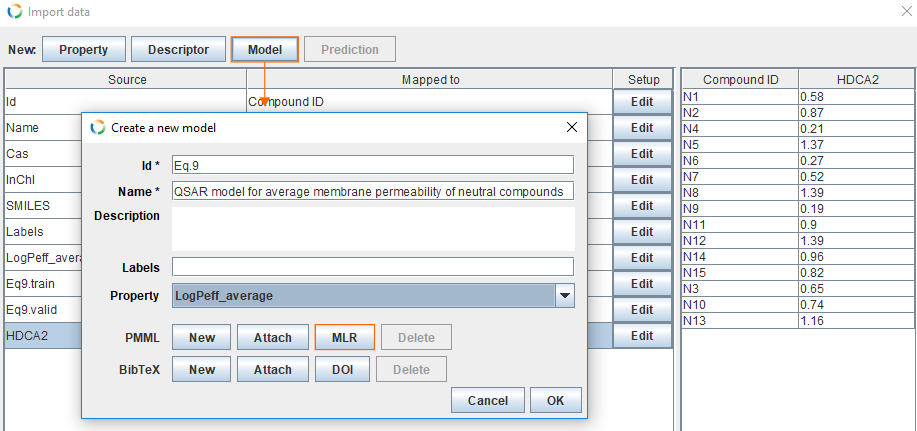
Add model
- To create a new model click Model, which opens the new model dialog.
- Fill Id field with Eq.9.
- For Name we use QSAR model for average membrane permeability of neutral compounds.
- For Property choose LogPeff_average.
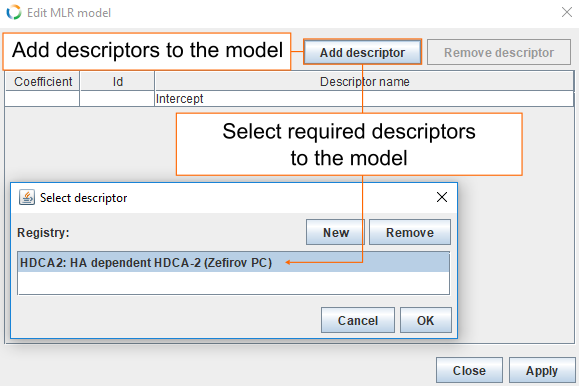
- Add PMML representation for the model by clicking MLR.
- In the MLR dialog click Add descriptor, select HDCA2 and click OK.
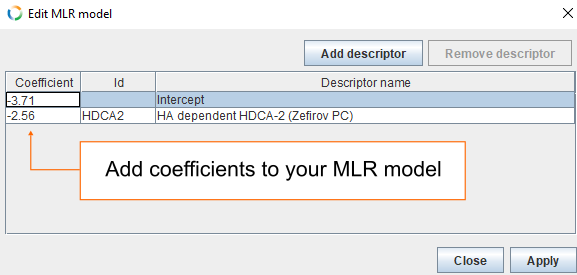
- Add coefficients for Intercept (-3.71) and for HDCA2 (-2.56) and click Apply.
- Finally, click OK.
Map predictions
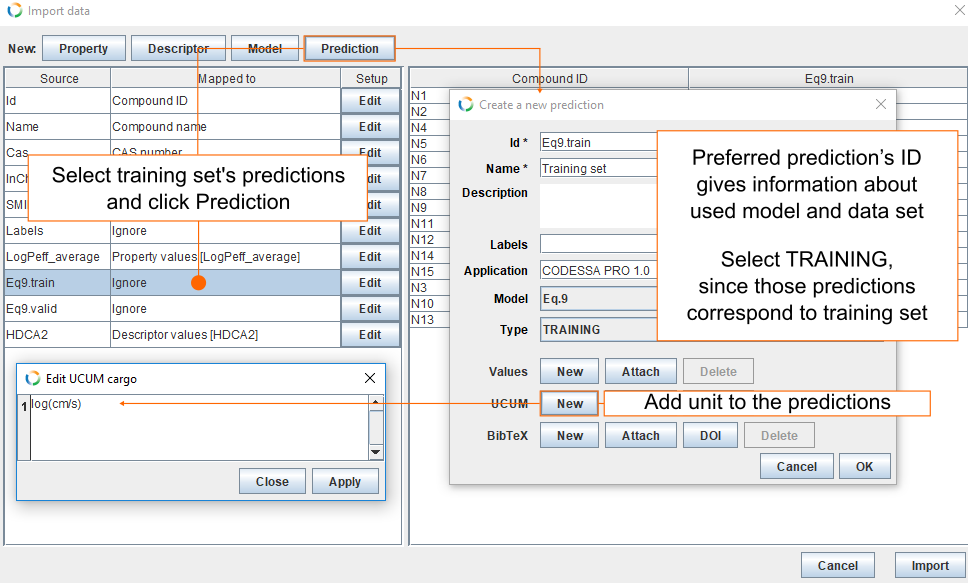
- Select Eq9.train and click Prediction. This opens a new dialog from where you can fill the fields for the new prediction.
- Preferred prediction's ID gives information about used model and used data set. By default Id field is filled with columns name (Eq9.train).
- For Name we use Training set.
- Modelling was done with CODESSA PRO 1.0 and this goes into Application field.
- For Model select Eq.9.
- For data set Type, select TRAINING.
- Click New in UCUM field and insert log(cm/s) and click Apply.
- Finally, click OK.
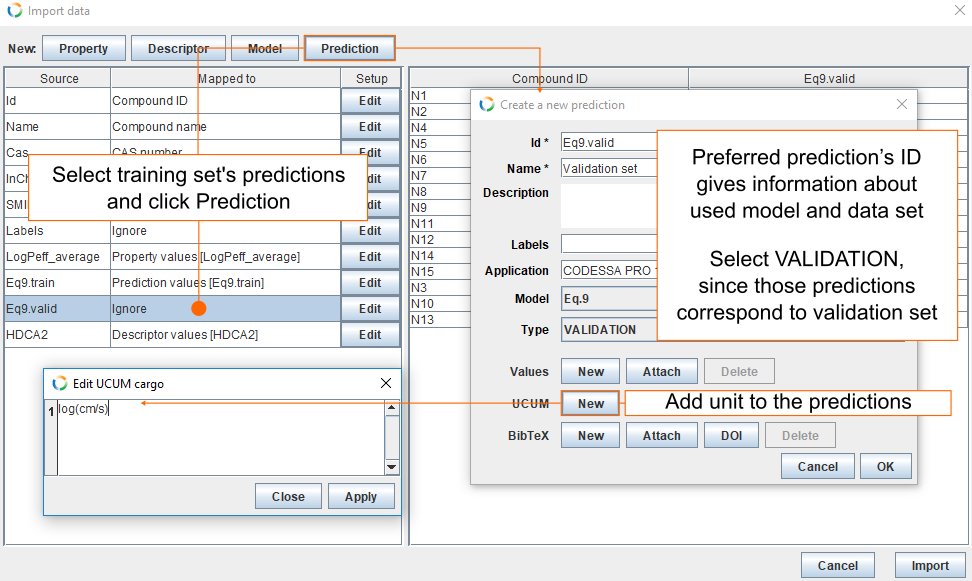
- Select Eq9.valid and click Prediction. This opens a new dialog from where you can fill the fields for new predictions.
- Preferred prediction's ID gives information about used model and used data set. By default Id field is filled with columns name (Eq9.valid).
- For Name we use Validation set.
- Modelling was done with CODESSA PRO 1.0 and this goes into Application field.
- For Model select Eq.9.
- For data set Type, select VALIDATION.
- Click New in UCUM field and insert log(cm/s) and click Apply.
- Click OK.
- Now we have mapped all the fields. Finally, click Import to add everything to the archive.
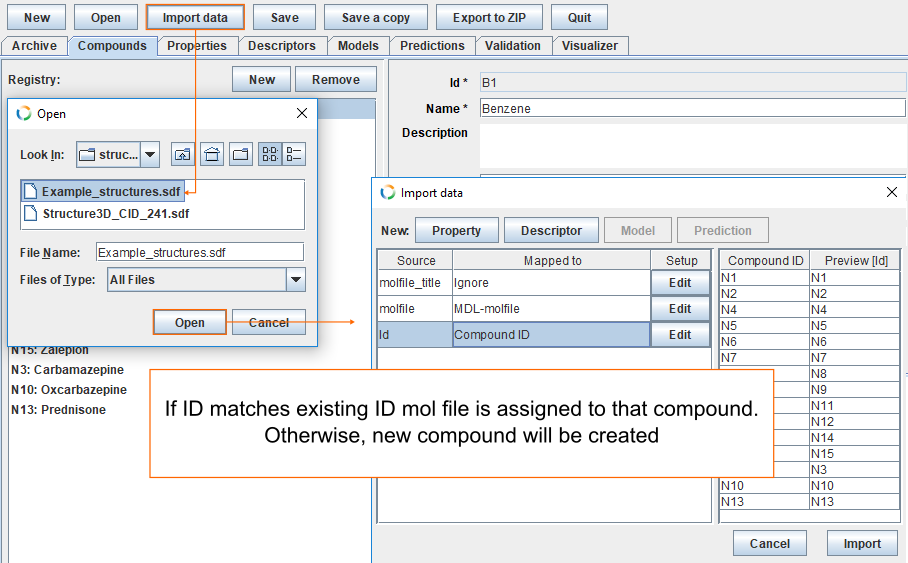
Import 3D-structures
To add mol-files to the archive, click Import data, find the file Example_structures.sdf and click Open. If ID matches existing ID, mol-file is assigned to that compound. Otherwise, new compound will be created.
Step 2: Add archive description Step 4: Validate created archive
