Chapter 2: QsarDB data format
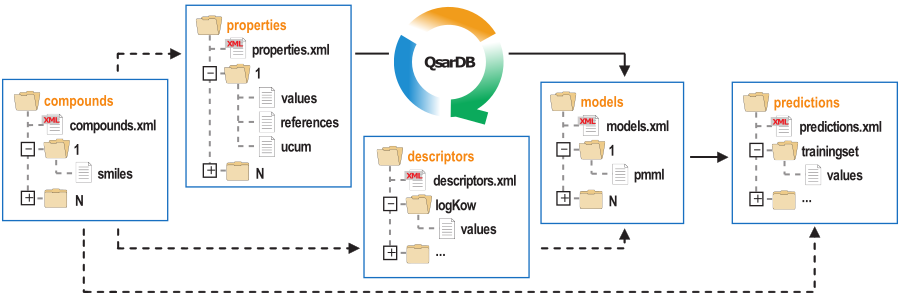
A more detailed description about QsarDB data format can be found in the respective scientific publication. In short, QsarDB data (see figure below) format is a systematic collection of folders and files that are linked together through strong (solid arrows) and weak (dashed arrows) relationships between the five main folders. The main folders are called containers and their names express the central components of QSAR analysis (compounds, properties, descriptors, models and predictions). Attributes for each component are in the XML-file and cargos (a document attachments) are in a separate folder.
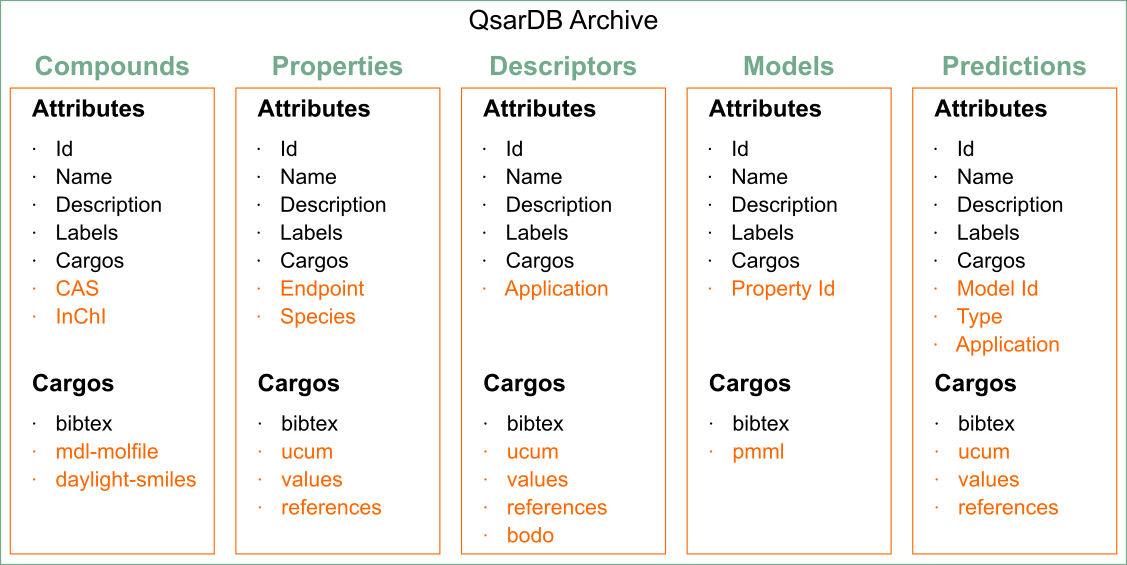
Some attributes and cargos for those five Containers are same for all, but depending on the container they can have some unique attributes and cargos (see figure below). For example, Property defines two attributes (i.e. Endpoint, Species). All the available attributes and cargos are explained in following paragraphs.
Archive
The path of the archive descriptor is “archive.xml”. It is not a Container, it rather characterizes the QDB archive as a whole and has two attributes.
Attributes
Name is a short description, i.e. one sentence. When the QDB archive is based on the research work already published in scientific literature then the name should correspond to the title of the article. If not published it is recommended to state the (i) endpoint, (ii) chemical class and (iii) statistical technique (for example: Linear regression model for the toxicity of anilines (pLC50) to algae (Pseudokircheriella subcapitata)).
Description is a longer description, i.e. one or more paragraphs. When the QDB archive is based on the research work already published in scientific literature then the description should correspond to the abstract of the article.
Compounds
Attributes
- Id is the symbolic identifier. Identifiers are used for forming relationships, which is why they must be unique within a container registry. For the consistency, the compounds ID should be the same as in the corresponding article.
- Name is a short description that provides complete and unambiguous identification of a Container object for a human agent. Recommended compounds Name is its preferred IUPAC name.
- Description is a longer description that summarizes all significant findings about particular compound. However, the description is not a place to discuss very detailed scientific hypotheses. The attribute must be in plain text.
- Labels is a list of text tokens. Every label (also known as a tag) defines a set with the same name.
- Cargos is a list of cargo identifiers. Every cargo identifier denotes a document attachment with the same name. There are two kinds of cargo identifiers:
- system cargos are part of the current QDB specification and employ simple one word identifiers
- extension cargo types must employ qualified identifiers.
- CAS CAS registry number (CAS RN) is the identifier for chemical substances that includes all categories of chemical compounds in the CAS registry database
- InChI IUPAC International Chemical Identifier (InChI) is a standardized textual identifier. The InChI value must be a standard InChI (starts with prefix “InChI = 1S”), non-standard InChI must be as structure Cargo
Cargos
- bibtex A BibTeX database consists of BibTeX data entries. A BibTeX data entry is a data structure which has a unique identifier and a number of publication type-dependent required and optional fields.
- mdl-molfile MOL file as structure Cargo (3D structure information)
- daylight-smiles Simplified Molecular-Input Line-Entry System (SMILES) string string as structure Cargo (2D structure information)
Properties
Attributes
- Id is the symbolic identifier. Property ID must be as close as possible to the ID used in article (e.g. logBCF, BCF_class) to guarantee consistency between two sources.
- Name is a short description. Property Name should give short description about property [e.g. Experimental logarithmic BCF; Experimental BCF class (nB - non-bio-accumulative, B - bioaccumulative)]
- Description is a longer description that summarizes all significant findings about the property.
- Labels is a list of text tokens.
- Cargos is a list of cargo identifiers.
- Endpoint is the experimental test classification, i.e. physico-chemical, biological, or environmental effect that has been measured. The current QDB specification uses the QMRF classification system (e.g. 2. Environmental fate parameters 2.4. Bioconcentration)
- Species is the name of the species according to the binomial nomenclature. This attribute is only applicable to Properties that represent biological activities. (e.g. Cyprinos carpio (common carp))
Cargos
- bibtex contains information from where experimental measurements originate.
- ucum The unified code for units of measure (UCUM) contains unit of the experimental property. If property is dimensionless UCUM can be omitted. Documentation for UCUM can be found in http://unitsofmeasure.org/ucum.html
- values property values are stored as tabular data in a text file. This file is formatted as a “tab-separated values” (TSV) document. The first column holds Compound identifiers and the second column holds activity/property values.
- references Bibliography references for individual property values are stored in a “tab-separated values” (TSV) document. The first column holds Compound identifiers and the second column holds target BibTeX data entry identifiers.
Descriptors
Attributes
- Id is the symbolic identifier. Preferred descriptor’s ID must ideally match descriptor calculation software’s internal descriptor ID.
- Name is a short description. Preferably must be same as in descriptor calculation software documentation.
- Description is a longer description that summarizes all significant findings about the descriptor.
- Labels is a list of text tokens.
- Cargos is a list of cargo identifiers.
- Application Application should refer to used software and its version (e.g. PaDEL-Descriptor 2.18)
Cargos
- bibtex contains information from where experimental measurements originate.
- ucum If descriptor has an unit it must be represented as UCUM http://unitsofmeasure.org/ucum.html
- values descriptor values are stored as tabular data in a text file. This file is formatted as a “tab-separated values” (TSV) document. The first column holds Compound identifiers and the second column holds descriptor values.
- references Bibliography references for individual descriptor values are stored in a “tab-separated values” (TSV) document. The first column holds Compound identifiers and the second column holds target BibTeX data entry identifiers.
- bodo Blue Obelisk Descriptor Ontology (BODO) is the ontology of cheminformatics algorithms. It is used primarily by the CDK and JOELib/JOELib2 cheminformatics libraries.
Models
Attributes
- Id is the symbolic identifier. Model’s ID must give enough information to precisely locate model in the article. (e.g. Tab1.Model1, Eq8).
- Name is a short description. It is recommend not including property’s name into the model’s name to avoid repetition (e.g. Melting point model). If certain group of chemicals were modelled, the name of the chemical class will add extra information for the model user (e.g. Model for hydrocarbons). Another example of model’s name is ‘Imbalanced model towards B-compounds’
- Description is a longer description that summarizes all significant findings about the model.
- Labels is a list of text tokens.
- Cargos is a list of cargo identifiers.
- PropertyId is the identifier of the Property that was used in the modelling.
Cargos
- bibtex contains information from where used model originate.
- pmml Predictive Model Markup Language (PMML) is an XML based data format for the representation of statistical and data mining models. Documentation and examples of use can be found at http://www.dmg.org/v4-1/GeneralStructure.html
Predictions
Attributes
- Id is the symbolic identifier. Preferred prediction’s ID gives information about used model and data set (e.g. M1.train, M2.valid)
- Name should give short description about data used for predictions (e.g. Training set, Validation set)
- Description is a longer description that summarizes all significant findings about the predictions.
- Labels is a list of text tokens.
- Cargos is a list of cargo identifiers.
- ModelId is identifier of the Model that was used for making predictions.
- Type Type can be “training”, “validation” or “testing”.
- training - Predictions for a data set used for the model development
- validation – Model benchmarking and making predictions on known chemical systems
- testing - Making predictions on unknown chemical systems
- Application refers to software and its version which was used for modelling (e.g. Random Forest 4.6-7)
Cargos
- bibtex contains information from where predictions originate.
- ucum If prediction has an unit it must be represented as UCUM http://unitsofmeasure.org/ucum.html
- values prediction values are stored as tabular data in a text file. This file is formatted as a “tab-separated values” (TSV) document. The first column holds Compound identifiers and the second column holds prediction values.
- references Bibliography references for individual prediction values are stored in a “tab-separated values” (TSV) document. The first column holds Compound identifiers and the second column holds target BibTeX data entry identifiers.
